Superbug genome successfully decoded
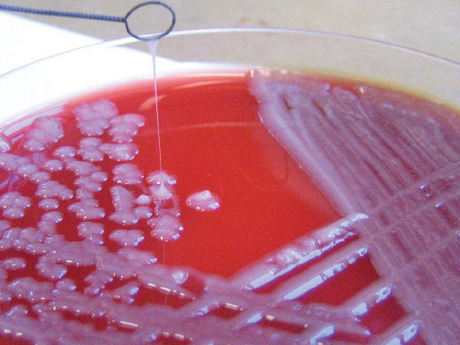
The University of Melbourne has led an international research collaboration to decode the genes of Klebsiella pneumoniae (KP), a bacterium found in hospitals throughout the developing and developed world.
Involving 37 research institutions around the world and bacterial samples from six countries, representing 300 strains, the study is the largest ever genetically decoded collection of this bacterial pathogen.
The scientific community does not have much information on the KP bacterium, whose drug-resistant strain (KPC) has caused significant problems in America and Europe for over a decade. The new study, published in the journal Proceedings of the National Academy of Sciences, is the first large-scale project that uses genomic technology to understand exactly what KP is and what it is capable of.
“KP is commonly found in the environment and can acquire resistance genes, become established in hospitals and then become a major health problem,” explained co-author Professor Dick Strugnell from the University of Melbourne’s Doherty Institute.
“The bacterium has evolved a thick sticky sugar coat which stops it drying out. This coat contributes to the formation of biofilms of the bacteria which are hard to remove from hospitals with traditional cleaning methods.”
Lead author Dr Kathryn Holt, of the University of Melbourne’s Bio21 Institute, said that in most cases KP infects people who are already weakened by illness, but there is a risk that it will evolve to become a significant threat to healthy individuals. Given the ease with which different strains of the bacteria can share genetic information, she is concerned that antibiotic resistance genes may soon appear in more infectious Klebsiella strains.
“So far we have been extremely lucky in that most of the antibiotic-resistant strains are not highly virulent to humans,” Dr Holt said. “Unfortunately, Klebsiella strains are very good at swapping around genes that encode antibiotic resistance, so it’s probably just a matter of time before we see this.
“The bacterium is really good at evolving through acquiring new genes. We looked at 300 different strains and every second one we looked at was completely new, so that tells you there’s a lot of diversity out there and we don’t really have a handle on this at all.”
The study provides a foundation for further research into how these bacteria adapt to new niches and become more diverse. The findings of the paper could also prove useful for designing vaccines to prevent, rather than treat, KP infections.
“The Microbiological Diagnostic Unit at the Doherty Institute, headed by Professor Ben Howden, is leading the work to track KPC,” said Dr Holt, “but more investment from the Victorian and the federal governments in research and surveillance for these drug-resistant organisms could go a long way.”
Personality influences the expression of our genes
An international research team has used artificial intelligence to show that our personalities...
Pig hearts kept alive outside the body for 24 hours
A major hurdle for human heart transplantation is the limited storage time of the donor heart...
Breakthrough antibiotic for mycobacterial infections
The antibiotic candidate, named COE-PNH2, has been optimised to target Mycobacterium...







